Research
Evolutionary Genetics and Genomics of Ecological Divergence
A central goal of evolutionary biology is to understand how diversity arises and is maintained in natural populations. We are taking an integrative approach that combines genomics, genetics and physiology to investigate the mechanisms of ecological divergence in the cosmopolitan, multicellular cyanobacterium Mastigocladus laminosus at multiple spatial scales. For example, at White Creek, a geothermally-influenced, nitrogen-limited stream in the Lower Geyser Basin of Yellowstone National Park (image at right), variation in temperature performance has evolved and is maintained in the population despite strong gene flow in most regions of the genome. Genome-wide analyses have revealed several loci that exhibit extreme genetic differentiation along the White Creek temperature gradient. Many are involved in the development and metabolism of the heterocyst, the oxygen-sensitive, nitrogen-fixing cell produced by this bacterium during nitrogen limitation. We are currently investigating the functional significance of a deletion polymorphism that impacts the gas permeability of the heterocyst's protective envelope. Spatial variation at White Creek appears to favor alternative alleles due to thermodynamic effects on physiological performance. Remarkably, this polymorphism arose early during Mastigocladus diversification, has persisted for millions of years and has spread throughout the organism’s range. This illustrates how spatially varying selection can contribute to both local adaptation and the distribution of biological diversity on a global scale.

Genome Evolution in Chlorophyll d-producing Cyanobacteria
Chlorophyll d-producing cyanobacteria (Acaryochloris) are a relatively recently discovered but biogeochemically important group of oxygenic photoautotrophs that are unique in their utilization of this far-red light absorbing, structural relative of Chl a as the major pigment in photosynthesis. As a result, these bacteria occupy a novel solar niche that is invisible to most other photosynthetic organisms. We have discovered and characterized a novel member of this group from the Salton Sea, a eutrophic saline lake in southern California. Genome sequencing of this and related strains has revealed that their genomes harbor an extraordinarily high number of gene duplicates for bacteria. As part of the UM NASA Astrobiology Institute, we seek to understand the contribution of gene duplication to environmental adaptation of these organisms and the role of paralogous copies of the recombination enzyme RecA for the unusual duplication dynamics of these genomes.
Adaptation at the Thermal Limit for Photoautotrophy
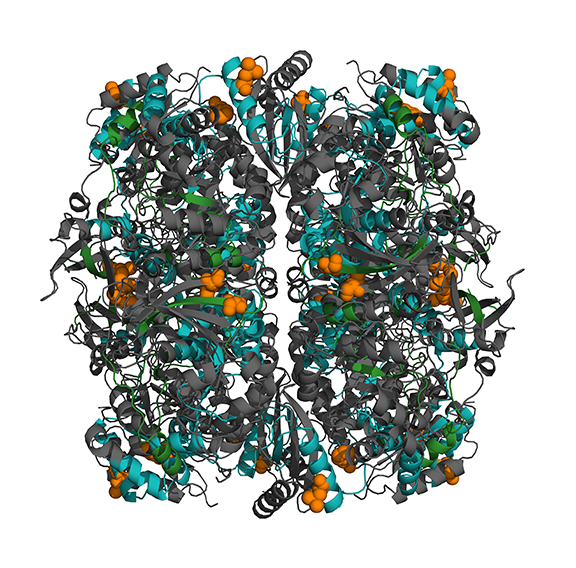
A long-standing question in evolutionary biology is how organisms adapt to novel environments. In North American hot springs, diversification of a clade of the cyanobacterium Synechococcus into hotter environments has resulted in the unique innovation of a light-driven ecosystem at temperatures up to 73 °C, the upper thermal limit for photosynthetic life. Closely related lineages of Synechococcus isolated from different locations along a hot spring temperature gradient differ in thermal ecology, and increasingly more thermotolerant Synechococcus lineages have diverged from less thermotolerant ancestors in response to directional selection for enhanced performance at higher temperature. We seek to understand the mechanisms of adaptive divergence during Synechococcus diversification, and whether adaptation to higher temperature has come with evolutionary costs for performance at lower temperatures. For example, we have recently taken a functional approach to show that the evolution of enhanced thermostability of the carbon fixation enzyme RuBisCO was essential for the innovation of a phototrophic lifestyle at temperatures greater than 70 °C. Contrary to the conventional view on the genetics of adaptation obtained from laboratory evolution experiments with microorganisms, RuBisCO adaptation involved the homologous recombination of existing genetic variation rather than the sequential fixation of novel beneficial mutations (in the image at right, stabilizing amino acids in the thermostable RuBisCO are in orange, and regions of the protein derived from recombination rather than vertical inheritance are colored green and blue, respectively).